

Developing techniques for multi-color, Live-cell single-molecule super resolution microscopy
In the laboratory for Biophysics at the Racah Institute of Physics, we develop and apply combinations of single molecule localization microscopy (SMLM), including photoactivated localization microscopy (PALM) and direct stochastic optical reconstruction (dSTORM) in multiple colors. These methods enable the study of signaling complexes in single molecule detail in intact (fixed and live) cells with resolution down to ~20nm. We further combine and synergize these techniques with related techniques, such as Super-resolution Optical Fluctuation Imaging (SOFI) and single particle tracking (SPT), for optimal live cell imaging and physical characterization at the single molecule level.
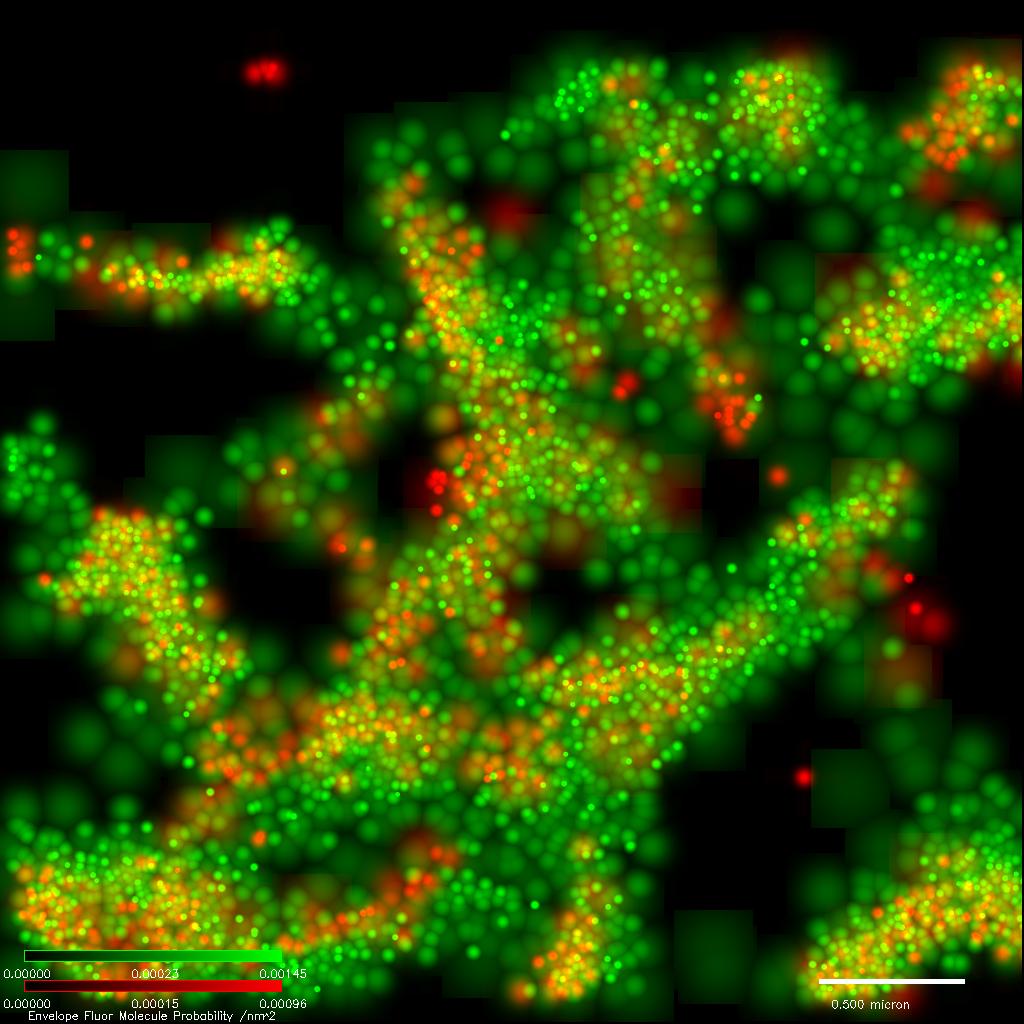
Single molecule super-resolution imaging of three molecular species in locations of high molecular densities has allowed us to study the complexity of molecular interactions, including potential cooperativity or competition in molecular binding.
We aim to develop a fundamental understanding of critical mechanisms of cell activation in health and disease in single molecule detail. Ultimately, such a level of understanding could serve to identify novel and efficient ways of intervening in aberrant signaling pathways and cellular malfunctions. To overcome current limitations in research techniques, we rely on cutting-edge microscopy techniques at the single molecule level of intact cells on functionalized interfaces, advanced statistical methods and quantifiable models based on physics of complex systems. Research in our lab is currently focused on the activation of T cells, which plays a central role in mounting adequate immune responses to foreign pathogens. Previous work has resulted in several intriguing findings, such as that signaling complexes at the plasma membrane of T cells have nanoscale structure and organization that facilitate intact cell activation.
The plasma membrane is a complex fluid where transmembrane proteins diffuse and interact to facilitate cell function. Membrane protein mobility is affected by multiple mechanisms, including crowding, trapping, medium elasticity and structure, thus limiting our ability to distinguish them in intact cells. In this study, we characterize the mobility and organization of transmembrane proteins at the plasma membrane of live T cells, using single particle tracking and photoactivated-localization microscopy. Using mobility characteristics, we segment individual trajectories into subpopulations with distinct Gaussian step-size distributions. In this way, we are able to separate mixed mechanisms of subdiffusion that act differentially on tracer patricles. Studying short transmembrane fragments of the (HIV1 coat) gp41 protein, we observe that particles of low-to-medium mobility consist of clusters, diffuse in a viscoelastic and fractal-like medium and are enriched at the center of the cell footprint. Particles of high mobility undergo weak confinement and are more evenly distributed. This study presents a methodological approach to resolve simultaneous mixed subdiffusion mechanisms acting on polydispersed samples and in complex media such as cell membranes.
Theme by Danetsoft and Danang Probo Sayekti inspired by Maksimer